Chemical ligation
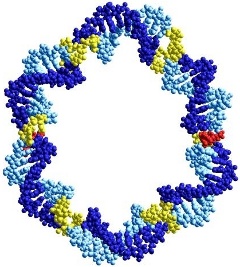 We are using the copper-catalysed alkyne-azide coupling reaction (click
chemistry) to synthesise large, covalently closed cyclic DNA structures and
catenanes for use as decoys for transcription factors, and as scaffolds in
nanotechnology.
We are using the copper-catalysed alkyne-azide coupling reaction (click
chemistry) to synthesise large, covalently closed cyclic DNA structures and
catenanes for use as decoys for transcription factors, and as scaffolds in
nanotechnology.
Our first paper in this field was one of the most cited JACS papers in 2007 (View paper) , and two other publications (View paper, View paper) were amongst the most accessed papers in ChemBioChem.
Collaborations with Prof. Bengt Norden’s group in Gothenburg involved the use of DNA click chemistry to elucidate the binding mechanism of a novel threading DNA intercalator (View paper) and led to the synthesis of addressable high-information-density DNA nanostructures (View paper). These are molecular scaffolds on which molecules can be attached at pre-determined locations with nanometre precision so that they can communicate in a controlled manner by energy or electron transfer.
We have also developed oligonucleotide ligation chemistry to synthesise DNA templates containing modified backbones which can be copied by DNA polymerases during PCR. This discovery (View paper) shows that the chemical synthesis of entire genes is feasible (View paper). One of my lectures on our work in this area can be seen on YouTube (View lecture). The field of nucleic acid click chemistry has been reviewed by us (View Paper) (View Paper).