New methods for RNA synthesis
RNA synthesis is less efficient than DNA synthesis, owing the problems caused by the presence of the 2'-hydroxyl group. Protection and deprotection of this group during solid-phase synthesis reduces coupling efficiency and leads to side-reactions. Consequently constructs longer than ~80 nucleotides in length (far smaller than most biologically active RNA molecules) are difficult to prepare in high purity. Alternative methods including transcription and enzymatic ligation prevent the site-specific incorporation of multiple modifications at sugars, bases and phosphates so have limited scope.
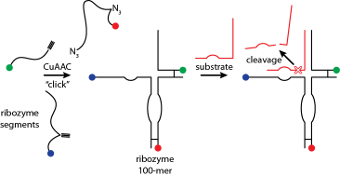 We have developed “click RNA ligation”
(View paper)
that overcomes the above limitations and allows the synthesis of RNA molecules
and DNA/RNA chimeras up to 100 nucleotides in length. We synthesize shorter
individual RNA oligonucleotides by automated solid phase synthesis, and use the
CuAAC reaction to ligate them. This reaction is efficient, orthogonal to
functional groups present in nucleic acids and works well in aqueous media. We
have used click RNA ligation to synthesize catalytically-active variants of the
hairpin ribozyme and the hammerhead ribozyme.
We have developed “click RNA ligation”
(View paper)
that overcomes the above limitations and allows the synthesis of RNA molecules
and DNA/RNA chimeras up to 100 nucleotides in length. We synthesize shorter
individual RNA oligonucleotides by automated solid phase synthesis, and use the
CuAAC reaction to ligate them. This reaction is efficient, orthogonal to
functional groups present in nucleic acids and works well in aqueous media. We
have used click RNA ligation to synthesize catalytically-active variants of the
hairpin ribozyme and the hammerhead ribozyme.
This chemistry could be applied to the synthesis of riboswitches, multivalent aptamers and components of ribosomes, tRNA and mRNA